I’ve put together this list of 10 pieces of free molecular biology software for Macs. I hope you will find at least some of it useful. If any of your favorite free programs are not included, please e-mail me and I’ll add them or you can leave a comment with a link. If you are a PC user, click here for the PC list.
1. Serial Cloner
Serial Cloner is a fantastic all-in-one workbench; import and manipulate sequences, construct plasmid and restriction site maps, determine %GC and fragment TM, extract and ligate fragments, perform virtual PCR… and lots more, all in one window using a very intuitive graphical interface.

2. 4Peaks
4Peaks is an extremely user-friendly DNA sequence chromatogram viewer and editor from the extremely talented guys at Mekentosj. It’s miles better than any of its clunky counterparts… try it, you’ll love it!
3. Beware of Molecules
This handy little application calculates the molecular weight of a molecule from its empirical formula. It has over a hundred preset compounds, which can be added to. Put that calculator away.
Free Dna Sequence Analysis Software
4. Papers
This one is not actually free, but is very cheap for the job it does. Papers is an application for retrieving, archiving, and viewing PDF files. From the Papers interface, Pubmed searches can be performed and PDF files, complete with all metadata can be downloaded. Another piece of brilliant software from Mekentosj and a bargain at only 29 euros ($40).
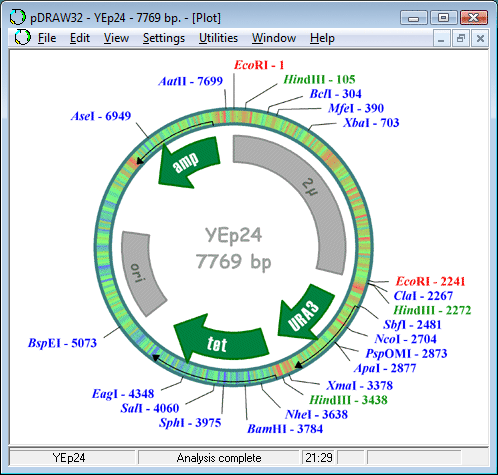
What is Sequencher for Mac. Sequencher is the industry standard software for DNA sequence analysis. It works with all automated sequencers and is widely known for its lightning-fast contig assembly, short learning curve, user-friendly editing tools, and superb technical support. First released almost 15 years ago, Sequencher is currently used. I would appreciate any suggestions on editing software (preferably a free one, but I'm curious about anything that is available). The main thing I'd like be able to do is to view trace files.
5. Geneious
Geneious is a software package of genome & proteome research tools for protein, DNA, or molecular visualization, literature searching and storage…and more. The Pro version does LOTS more.
Dna Sequence Analysis Software
- Geneious Prime is a powerful bioinformatics software solution packed with fundamental molecular biology and sequence analysis tools. Perform a wide-range of cloning and primer design operations within one interface. Take charge with industry-leading assembly and mapping algorithms, and superior visualizations.
- Art's Free Software Page (List of free software that is relevant to the Molecular Biology and Structural Biology fields. For different platforms: mac-windows-unix) General Software. Tucows Mac Software. Enzyme X A full featured macos restriction cutting analysis software.
6. PDF skim
I’m cheating here slightly because I have mentioned PDF skim in another article recently, but it can’t be left out of this list. PDF skim allows you to annotate a PDF file with your own notes and the makers hope that it will reduce the need for printing out PDF files. Save the rainforests – skim your PDFs.
7. Lab Assistant
I’m sorry, this is yet another application from Mekentosj. Actually, I’m not sorry because this is yet another GREAT piece of software from these guys. Lab Assistant helps you organize your experiments with sticky “to-do” lists, timers, a calendar, and a lab journal. No scientist should be without this!
8. ApE
ApE is an all-in-one plasmid and sequence workbench. Sequences can be uploaded to ApE manually, direct from the NCBI database, or from ABI chromatogram traces. ApE can be used for sequence annotation, restriction mapping, primer design, and sequence alignment. A great all-round tool.
9. Cn3D
Cn3D… say it out loud… “see in 3D”. This great piece of software from NCBI is a sequence viewer with a difference. Not only does it perform 2D alignments, but it also allows the user to see the position residues in the 3D protein sequence. Great for mutagenesis studies.
10. BioToolKit
This is one packed toolkit! Primer design, antibody design, microarray analysis, calculators for molecular weight, molar concentration and OD, centrifugation speed converter, label printing templates… and much more! Phew!